Methodological manual "Statistical analysis and visualization of data using R." Methodological manual "Statistical analysis and visualization of data using R" Statistical analysis and
Course program
Elements of programming in R
- Descriptive Statistics and Visualization
- For example, what is more important: the average check or the typical check?
Cluster analysis
- What problem is being solved? Divide a group of objects into subgroups.
- Example task. Segmentation of sites, identification of similar sites.
- Methods studied. Hierarchical cluster analysis, k-means method, K-medoid method.
Testing statistical hypotheses
- What problem is being solved? Compare two groups of objects.
- Example task. A/B testing of user behavior on different versions site pages.
- Methods studied. Test for proportions, Student's t test, Livigne test, Wilcoxon-Mann-Whitney test
Linear regression analysis.
- Example task. Estimate how much prices for used cars fell after the increase in customs duties.
- Methods studied. Variable selection, collinearity, influential observations, analysis of residuals. Nonparametric regression (kernel smoothing). Forecasting short series with a seasonal component using linear regression
Forecasting
- What problem is being solved? Build a time series forecast
- Example task. Predict website traffic for 6 months in advance.
- Method being studied. Exponential smoothing
Machine Learning (Pattern Recognition)
- Example task. Recognize the gender and age of each site visitor
- Methods studied. K-nearest neighbor Classification Trees (CART) method. Random forests. Gradient boosting machine
Course grades
Listeners will be given 14 laboratory work. The course is graded according to the following rule:
- Excellent - all work has been accepted;
- Good - all the works have been accepted, except one?;
- Satisfactory – all works except two are accepted;
- Unsatisfactory - in other cases.
The laboratory work is that
- the listener is given a data set and a question;
- the listener answers the question, supporting his statements with tables, graphs and a script written in the R language;
- The listener answers additional questions.
Sample question. Suggest parameters that will ensure optimal operation of the Random Forest algorithm when recognizing a wine brand based on the results of chemical analysis.
What you need to know to take the course
It is assumed that course participants have already taken a course in probability theory.
Literature
- Shipunov, Baldin, Volkova, Korobeinikov, Nazarova, Petrov, Sufiyanov Visual statistics. Using R
- Masticsky, Shitikov Statistical analysis and data visualization using R
- Bishop Pattern Recognition and Machine Learning.
- James, Witten, Hastie, Tibshirani. An Introduction to Statistical Learning. With Applications in R.
- Hastie, Tibshirani, Friedman. The Elements of Statistical Learning_Data Mining, Inference, and Prediction 2+ed
- Crawley. The R Book.
- Kabacoff R in Action. Data analysis and graphics with R.
Teachers
List of lectures
Introduction to R: Basic Commands. Median, quantiles and quartiles. Bar chart. Bar chart. Pie chart. Scatter diagram. Scatterplot matrix. Use of color in graphics. Boxes with whiskers (box diagram). Typical sample observation: arithmetic mean, median, or trimmed mean. Choosing a way to describe a typical value that is adequate to the analyzed data. Lognormal distribution. Outliers and extreme observations.
Hierarchical cluster analysis. Cluster, distances between objects, distances between clusters. Algorithm for constructing a dendrogram. Scree/elbow. Data standardization. Typical mistakes when preparing data. Interpretation of results.
K-means method. Random number sensors, sensor grain. Visualization of the k-means method algorithm. Methods for determining the number of clusters. NbClust library. Scree/elbow. Multidimensional scaling for cluster visualization.
Testing statistical hypotheses. Hypotheses of agreement, homogeneity, independence, hypotheses about distribution parameters.
Testing statistical hypotheses. Type I and type II errors, p-value and significance level, statistical hypothesis testing algorithm and interpretation of results. Hypothesis of normal distribution. Shapiro-Wilk and Kolmogorov-Smirnov tests. Minor deviations from normality. Comparison of samples. Independent and paired samples. Choosing between Student's t-test, Mann-Whitney-Wilcoxon test and Mood test. Varieties of Student's t-tests and comparison of variances. Visualization for comparisons. One-sided and two-sided tests.
Testing statistical hypotheses. Comparison of samples. Independent and paired samples. Choosing between Student's t-test, Mann-Whitney-Wilcoxon test and Mood test. Varieties of Student's t-tests and comparison of variances. Visualization for comparisons. One-sided and two-sided tests. Independence. Pearson, Kendall and Spearman correlation coefficients, typical mistakes when studying the connection between two phenomena. Visual inspection of findings.
Linear regression analysis Model, interpretation of coefficient estimates, multiple coefficient of determination. Interpretation of the multiple coefficient of determination, restrictions on the scope of its application. Identification of the most significant predictors and assessment of the contribution of each predictor. Algorithms for adjusting the constructed models. Collinearity.
Linear regression analysis: forecasting short time series.
Forecasting based on a regression model with seasonal indicator (dummy, structural) variables. Trend, seasonal components, changes in the nature of the series, outliers. Logarithmization is a technique for converting multiplicative seasonality into additive seasonality. Indicator variables. Retraining.
Linear regression - analysis of residuals. Violations of model restrictions of the Gauss-Markov theorem. Residue analysis. Specification error. Multicollinearity, Tolerance and VIF. Checking the constancy of variances of residuals. Correction of models in the presence of deviations in the distribution of residuals from normality. Cook's distance and leverage. Durbin-Watson statistics. Reducing the number of seasonal adjustments.
Exponential smoothing Holt-Winters method. Local trend, local seasonality.
Terminology: Machine Learning, Artificial Intelligence, Data Mining and Pattern Recognition.
K-nearest neighbor method. The consistency of the method. Lazy learning (lazy learning). Feature Selection. Cross-validation. k-fold cross-validation. Overfitting. Training and test samples.
K-nearest neighbor method Examples. Determining the number of nearest neighbors. Contingency table for determining the quality of the method.
CART classification trees. Geometric representation. Representation as a set of logical rules. Tree view. Nodes, parents and children, leaf nodes. Threshold values. rpart library. Impurity measures. Methods for measuring purity: Gini, entropy, classification errors. Tree learning rules. Library rpart.plot.
The blog "R: Data Analysis and Visualization" has existed for more than three and a half years. A few months ago, the idea was born to summarize the methodological messages published here over all this time in the form of an e-book. The author of the idea, and subsequently the co-author of the book, was Doctor of Biological Sciences Vladimir Kirillovich Shitikov (). We are pleased to present you the result as our New Year's gift.
Toolkit " Statistical analysis and data visualization using R"is addressed primarily to students, graduate students, young and established scientists, as well as professional analysts who have had no previous experience working with R. Following the traditions of the blog, we tried, if possible, to avoid the abuse of “ritual” phrases characteristic of numerous manuals on applied statistics , quoting well-known theorems and presenting multi-level calculation formulas, the emphasis was placed, first of all, on practical application - on the fact that the reader, guided by what he read, could analyze his data and present the results to colleagues.
- Chapter 1: Basic Components of the R Statistical Environment
- Chapter 2: Description of the R language
- Chapter 3: Basic R Graphics Features
- Chapter 4: Descriptive Statistics and Fitting Distributions
- Chapter 5: Classical methods and criteria of statistics
- Chapter 6: Linear Models in Analysis of Variance
- Chapter 7: Regression models of relationships between quantitative variables
- Chapter 8: Generalized, Structural and Other Regression Models
- Chapter 9: Spatial Analysis and Cartogram Creation
The official current version of the book in PDF format (~11 MB) is available for free download from two sites:
- GitHub repository: https://github.com/ranalytics/r-tutorials
- Website of the Institute of Ecology of the Volga Basin RAS: http://www.ievbras.ru/ecostat/Kiril/R/
On the same two resources you can find the R code scripts and data sets needed to reproduce the examples discussed in the book.
We will be grateful for any of your comments and suggestions regarding this work - please send them to e-mail rtutorialsbook["dog"]gmail.com
As noted above, the book is distributed completely free of charge. However, if you find it useful and you find it appropriate to thank the authors for their work, you can transfer any amount using the following button (all transactions are carried out in safe mode through an electronic payment system
"STATISTICAL ANALYSIS AND VISUALIZATION OF DATA USING R grass roots fruit foliage Heidelberg - London - Tolyatti 2014, ..."
-- [ Page 1 ] --
S.E. Mastitsky, V.K. Shitikov
STATISTICAL ANALYSIS AND
VISUALIZING DATA WITH R
grass roots fruits foliage
Heidelberg – London – Tolyatti
2014, Sergey Eduardovich Mastitsky, Vladimir Kirillovich Shitikov
Website: http://r-analytics.blogspot.com
This work is distributed under the license
Creative Commons Attribution - Non-Commercial
use – Under the same conditions 4.0 Worldwide.” Under this license, you may freely copy, distribute and modify this work, provided that you accurately credit its authors and source. If you modify this work or use it in your own works, you may distribute the result only under the same or a similar license. It is prohibited to use this work for commercial purposes without permission from the authors. For more information about the license, visit www.creativecommons.com
Please cite this book as follows:
Mastitsky S.E., Shitikov V.K. (2014) Statistical analysis and data visualization using R.
– EBook, access address:
http://r-analytics.blogspot.com
PREFACE 5
1. MAIN COMPONENTS OF THE R 8 STATISTICAL ENVIRONMENT
1.1. History of origin and basic principles of organization 8 of the R environment
1.2. Working with the R 11 interface command console
1.3. Working with R Commander 13 menus
1.4. Objects, packages, functions, devices 17
2. LANGUAGE DESCRIPTION R 23
2.1. R 23 data types
2.2. Vectors and matrices 24
2.3. Factors 29
2.4. Lists and tables 31
2.5. Importing data into R 37
2.6. Date and time representation; time series 40
2.7. Organization of calculations: functions, branches, loops 46
2.8. Vectorized calculations in R using apply-50 functions
3. BASIC GRAPHICS CAPABILITIES R 58
3.1. Scatterplots plot() and graphing options 58 functions
3.2. Histograms, Kernel Density Functions, and the 66 cdplot() function
3.3. Range diagrams 74
3.4. Pie and Bar Charts 77
3.5. Cleveland diagrams and one-dimensional scatter plots 84
4. DESCRIPTIVE STATISTICS AND FIT 97
DISTRIBUTION– – –
PREFACE
One of the main tools for understanding the world is the processing of data received by a person from various sources. The essence of modern statistical analysis is an interactive process consisting of exploration, visualization and interpretation of incoming information flows.The history of the last 50 years is also the history of the development of data analysis technology.
One of the authors fondly recalls the end of the 60s and his first program for calculating pair correlation, which was typed with metal pins on the “operating field” of 150 cells of the personal computer “Promin-2” weighing more than 200 kg.
Nowadays, high-performance computers and affordable software make it possible to implement the full cycle of the information technology process, which generally consists of the following steps:
° access to processed data (downloading them from different sources and compiling a set of interrelated source tables);
° editing loaded indicators (replacing or removing missing values, converting characteristics into a more convenient form);
° annotating data (to remember what each piece of data represents);
° receiving general information about the structure of the data (calculation of descriptive statistics in order to characterize the analyzed indicators);
° graphical presentation of data and calculation results in a clear, informative form (one picture is actually sometimes worth a thousand words);
° data modeling (finding dependencies and testing statistical hypotheses);
° presentation of results (preparation of tables and diagrams of acceptable publication quality).
In conditions when there are dozens of packages available to the user application programs, the problem of choice is relevant (sometimes tragic, if we recall “Buridan’s donkey”): which data analysis software should be preferred for your practical work? Here, the specifics of the problem being solved, the efficiency of setting up processing algorithms, the costs of purchasing programs, as well as the tastes and personal preferences of the analyst are usually taken into account. At the same time, for example, the template Statistica with its mechanical set of menu buttons cannot always satisfy a creative researcher who prefers to independently control the progress of the computational process. Combine Various types analysis, have access to intermediate results, manage the style of data display, add your own extensions of software modules and draw up final reports in the required form allow commercial computing systems, including high-level command language tools such as Matlab, SPSS, etc. An excellent alternative to them is the free software environment R, which is a modern and constantly evolving general-purpose statistical platform.
Today, R is the undisputed leader among freely distributed statistical analysis systems, as evidenced, for example, by the fact that in 2010 the R system won the annual open software competition Bossie Awards in several categories. The world's leading universities, analysts from major companies and research centers constantly use R when carrying out scientific and technical calculations and creating large information projects. The widespread teaching of statistics based on packages of this environment and the full support of the scientific community have led to the fact that the reduction of R scripts is gradually becoming a generally accepted “standard” both in journal publications and in informal communication between scientists around the world.
The main obstacle for Russian speaking users One thing that is certainly true about mastering R is that almost all the documentation for this environment exists on English language. Only since 2008, through the efforts of A.V. Shipunova, E.M. Baldina, S.V. Petrova, I.S. Zaryadova, A.G. Bukhovets and other enthusiasts, methodological manuals and books appeared in Russian (links to them can be found in the list of references at the end of this book; there are also links to educational resources, the authors of which make a feasible contribution to the promotion of R among Russian-speaking users).
This manual summarizes a set of methodological messages published by one of the authors since 2011 in the blog “R: Data Analysis and Visualization”
(http://r-analytics.blogspot.com). It seemed to us advisable to present all this somewhat disjointed material in a concentrated form for the convenience of readers, as well as expand some sections for completeness of presentation.
The first three chapters provide detailed instructions for working with R's interactive components, a detailed description of the language and the basic graphical capabilities of the environment.
This part of the book is quite accessible to those new to programming, although readers already familiar with the R language may find interesting code snippets there or use the descriptions of graphical options provided as a reference.
The following chapters (4-8) provide a description of common procedures for processing data and building statistical models, which is illustrated with several dozen examples. These include short description analysis algorithms, the main results obtained and their possible interpretation. We tried, if possible, to avoid the abuse of “ritual” phrases typical of numerous manuals on applied statistics, citing well-known theorems and citing multi-story calculation formulas. The emphasis was, first of all, on practical application - so that the reader, guided by what he had read, could analyze his data and present the results to colleagues.
The sections of this part are built according to the complexity of the presented material.
Chapters 4 and 5 are aimed at the reader with an interest in statistics only as part of an introductory university course. Chapters 6 and 7, within the framework of the unified theory of general linear models, present variance and regression analyzes and provide various algorithms for the study and structural identification of models. Chapter 8 is devoted to some modern methods for constructing and analyzing generalized regression models.
Since spatial analysis and the display of results on geographic maps and diagrams are of constant interest to researchers, Chapter 9 provides some examples of such visualization techniques.
We address our manual to students, graduate students, as well as young and established scientists who want to master data analysis and visualization using the R environment. We hope that by the end of reading this manual you will have some understanding of how R works, where you can get further information, as well as how to cope with simple and quite complex data analysis tasks.
Files with R code scripts for all chapters of the book, as well as the necessary source data tables for their execution, are freely available for download from the GitHub repository https://github.com/ranalytics/r-tutorials, as well as from the website of the Institute of Ecology of the Volga Basin RAS link http://www.ievbras.ru/ecostat/Kiril/R/Scripts.zip.
It should be noted that the text in this manual is presented in the author's edition and therefore, despite all our efforts, there is a possibility that it may contain typos, grammatical inaccuracies and unfortunate phrases. We will be grateful to you, Reader, for reporting these, as well as other defects found, by e-mail [email protected]. We will also be grateful for any other comments and suggestions you may have regarding this work.
– – –
1. MAIN COMPONENTS OF THE STATISTICAL ENVIRONMENT R
1.1. History of origin and basic principles of organization of the R environment The system of statistical analysis and data visualization R consists of the following main parts:
° high-level programming language R, which allows one line to implement various operations with objects, vectors, matrices, lists, etc.;
° a large set of data processing functions collected in separate packages;
° a developed support system, including updating environment components, interactive help and various educational resources designed both for the initial study of R and subsequent consultations on emerging difficulties.
The beginning of the journey dates back to 1993, when two young New Zealand scientists Ross Ihaka and Robert Gentleman announced their new development, which they called R. They took the programming language of a developed commercial system as a basis statistical processing S-PLUS data and created its free, free implementation, which differs from its progenitor in its easily expandable modular architecture. Soon, a distributed system for storing and distributing packages for R arose, known by the abbreviation "CRAN" (Comprehensive R Archive Network - http://cran.r-project.org), the main idea of the organization of which is constant expansion, collective testing and operational distribution of applied data processing tools.
It turned out that such a product of continuous and well-coordinated efforts of the powerful “collective intelligence” of thousands of selfless intellectual developers turned out to be much more effective than commercial statistical programs, the cost of a license for which can be several thousand dollars. Since R is the favorite language of professional statisticians, all the latest advances in statistical science are quickly becoming available to R users around the world in the form of additional libraries. No commercial statistical analysis system is developing so quickly today. R has a large army of users who inform the authors of additional libraries and the R system itself about detected errors, which are promptly corrected.
The R calculation language, although it requires some effort to master, remarkable search skills and an encyclopedic memory, allows you to quickly perform calculations that are practically “as inexhaustible as an atom” in their diversity. As of July 2014, enthusiasts from all over the world have written 6,739 additional libraries for R, including 137,506 functions (see
http://www.rdocumentation.org), which significantly expand the basic capabilities of the system. It is very difficult to imagine any class of statistical methods that is not already implemented today in the form of R packages, including, of course, the entire “gentleman's set”: linear and generalized linear models, nonlinear regression models, experimental design, time series analysis, classical parametric and nonparametric tests, Bayesian statistics, cluster analysis and smoothing methods. With the help of powerful visualization tools, the results of the analysis can be summarized in the form of various graphs and charts. In addition to traditional statistics, the developed functionality includes a large set of algorithms for numerical mathematics, optimization methods, solving differential equations, pattern recognition, etc. Geneticists and sociologists, linguists and psychologists, chemists and doctors, and specialists in GIS and Web technologies.
The “proprietary” documentation for R is very voluminous and is not always well written (in the strange tradition of English-language literature, too many words are spent describing trivial truths, while important points are covered quickly). However, in addition to this, the world's leading publishers (Springer, Cambridge University Press and Chapman & Hall / CRC) or simply individual teams of enthusiasts have published a huge number of books describing various aspects of data analysis in R (see, for example, the list of references on the website "Encyclopedia of Psychodiagnostics", http://psylab.info/R:Literature). In addition, there are several active international and Russian R user forums where anyone can ask for help with a problem. In the bibliography, we list a couple of hundred books and Internet links that we advise you to pay special attention to while learning R.
Direct training practical work in R consists of a) mastering the constructs of the R language and becoming familiar with the features of calling functions that perform data analysis, and b) acquiring skills in working with programs that implement specific methods of data analysis and visualization.
The choice of R user interface tools is controversial and highly dependent on user tastes. Even authoritative experts do not have a consensus.
Some believe that there is nothing better than the standard R console interface. Others believe that for comfortable work It is worth installing any of the available integrated development environments (IDEs) with a rich set of button menus. For example, the free RStudio integrated development environment is a great option.
Below we will focus on the description of the console version and working with R Commander, but a review can help the reader’s further research different versions IDE, presented in the appendix to the book by Shipunov et al. (2014).
One R expert, Joseph Rickert, believes that the process of learning R can be divided into the following stages (for more details, see:
his article on inside-r.org):
1. Getting to know general principles the culture of the R community and the software environment in which the R language was developed and operates. Visit the main and auxiliary resources and master a good introductory textbook. Installing R on the user's computer and executing the first test scripts.
2. Reading data from standard operating system files and confidently using R functions to perform a limited set of statistical analysis procedures familiar to the user.
3.Usage basic structures R language for writing simple programs.
Writing your own functions. Familiarization with the data structures that R can work with and the more complex features of the language. Working with databases, web pages and external sources data.
4. Writing complex programs in the R language. Independent development and deep understanding of the structure of objects of the so-called S3- and S4-classes.
5. Development professional programs in the R language. Independent creation of additional library modules for R.
Most average R users stop at stage 3 because...
The knowledge acquired by this time is quite sufficient for them to perform statistical tasks in the profile of their main professional activity.
This is approximately the extent of our description of the R language within the framework of this manual.
Installing and configuring the basic R statistical environment is quite simple. As of July 2014, the current version is R 3.1.1 for 32 and 64-bit Windows (distributions for all other common operating systems are also available). You can download the system distribution kit along with a basic set of 29 packages (54 megabytes) completely free of charge from the main project website http://cran.r-project.org or the Russian “mirror” http://cran.gis-lab.info. The process of installing the system from the downloaded distribution does not cause any difficulties and does not require any special comments.
For the convenience of storing scripts, source data and calculation results, it is worth selecting a special working directory on the user’s computer. It is highly undesirable to use Cyrillic characters in the name of the working directory.
It is advisable to place the path to the working directory and some other settings options by changing any text editor system file C:\Program Files\R\Retc\Rprofile.site (it may have a different location on your computer). In the example below, the modified rows are marked in green.
In addition to specifying the working directory, these lines specify a link to the Russian source for downloading R packages and automatically launching R Commander.
Listing of the Rprofile.site file # Everything that follows the comment symbol "#" is ignored by the environment # options(papersize="a4") # options(editor="notepad") # options(pager="internal") # set display type reference information# options(help_type="text") options(help_type="html") # set local library location #.Library.site - file.path(chartr("\\", "/", R.home()) , "site-library") # When loading the environment, launch the R Commander menu # Add "#" signs if launching Rcmdr is not needed local(( old - getOption("defaultPackages") options(defaultPackages = c(old, "Rcmdr") ) )) # Define a mirror CRAN local((r - getOption("repos") r["CRAN"] - "http://cran.gis-lab" options(repos=r))) # Define the path to the working directory (any other one on your computer) setwd("D:/R/Process/Resampling") As for a “good introductory textbook,” any of our recommendations will be subjective. However, it is worth mentioning the officially recognized introduction to R by W. Venables and D. Smith (Venables, Smith, 2014) and the book by R. Kabakov (Kabaco, 2011), partly also because their Russian translation is available. Let us also note the traditional “manual for dummies” (Meys, Vries, 2012) and the manual (Lam, 2010), written with enviable Dutch pedantry. Of the Russian-language introductory courses, the most complete are the books by I. Zaryadov (2010a) and A. Shipunov et al. (2014).
1.2. Working with the R Interface Command Console The R statistical environment executes any set of meaningful R language instructions contained in a script file or represented as a sequence of commands issued from the console. Working with the console may seem difficult for modern users accustomed to push-button menus, since it is necessary to remember the syntax of individual commands. However, after acquiring some skills, it turns out that many data processing procedures can be performed faster and with less difficulty than, say, in the same Statistica package.
The R console is a dialog box in which the user enters commands and sees the results of their execution. This window appears immediately when the environment starts (for example, after clicking on the R shortcut on the desktop). In addition, the standard R graphical user interface (RGui) includes a script editing window and pop-up windows with graphical information (pictures, diagrams, etc.)
In command mode, R can work, for example, like a regular calculator:
To the right of the prompt symbol, the user can enter an arbitrary arithmetic expression, press the Enter key, and immediately get the result.
For example, in the second command in the picture above, we used the factorial and sine functions, as well as the built-in p. The results obtained in text form can be selected with the mouse and copied via the clipboard to any text file in the operating system (for example, a Word document).
When working with RGui, we recommend in all cases creating a file with a script (i.e. a sequence of R language commands that perform certain actions). As a rule, this is a regular text file with any name (but, for definiteness, it is better with the *.r extension), which can be created and edited with a regular editor like Notepad. If this file exists, it is best to place it in the working directory, and then after starting R and selecting the "File Open Script" menu item, the contents of this file will appear in the "R Editor" window. You can execute a sequence of script commands from the "Edit Run All" menu item.
You can also select with the mouse a meaningful fragment from any place in the prepared script (from the name of one variable to the entire content) and launch this block for execution. This can be done with four possible ways: from the main and context menu, the Ctrl+R key combination or a button on the toolbar.
In the presented figure, the following actions were performed:
° the R-object gadm with data on the territorial division of the Republic of Belarus was downloaded from the free Internet source Global Administrative Areas (GADM);
° Latinized city names are replaced with commonly used equivalents;
° using the spplot() function of the sp package, an administrative map of the republic is displayed in the graphic window, which can be copied to the clipboard using the menu or saved as a standard meta- or raster graphic file.
We will look at the meaning of individual operators in more detail in subsequent sections, but here we note that by selecting the Regions@data combination of symbols in the script and running the symbol combination Regions@data, we will receive the entire set in the console window data by object and a command made up of the selected gadm symbols, Regions@data$NAME_1, will give us a list of names of administrative centers before and after its modification.
Thus, the R Editor makes it easy to navigate through a script, edit and execute any combination of commands, and find and replace specific parts of code. The RStudio add-on mentioned above allows you to additionally perform code syntax highlighting, automatic code completion, “packaging” a command sequence into functions for their subsequent use, working with Sweave or TeX documents and other operations that will be useful to an advanced user.
R has extensive built-in reference material that can be accessed directly from RGui.
If you issue the help.start() command from the console, a page will open in your Internet browser that gives access to all help resources: main manuals, author's materials, answers to common questions, lists of changes, links to help on other R objects, etc. .d.:
Help for specific functions can be obtained using the following commands:
° help("foo") or? foo – help on the foo function (quotes are optional);
° help.search("foo") or ?? foo – search for all help files containing foo;
° example("foo") – examples of using the foo function;
° RSiteSearch("foo") – search for links in online manuals and mailing archives;
° apropos("foo", mode="function") – list of all functions with the combination foo;
° vignette("foo") – list of tutorials on the topic foo.
1.3. Working with the menu of the R Commander package A convenient tool for mastering calculations in R for a novice user is R Commander - platform-independent GUI in the style of a button menu, implemented in the Rcmdr package. It allows you to carry out a large set of statistical analysis procedures without resorting to preliminary memorization of functions in the command language, but it involuntarily contributes to this, since it displays all executed instructions in a special window.
You can install Rcmdr, like any other extensions, from the R console menu "Packages Install package", but it is better to run the command:
install.packages("Rcmdr", dependencies=TRUE) where enabling the dependencies option will ensure that the full set of other packages that may be required when processing data through the Rcmdr menu is installed.
R Commander is launched when the Rcmdr package is loaded through the "Packages Enable Package" menu or with the library(Rcmdr) command. If for some reason it was decided to analyze the data exclusively using R Commander, then for automatic download this graphical shell When starting R, you need to edit the Rprofile.site file as shown in section 1.1.
Let's consider working in R Commander using the example of correlation analysis of data on the level of infection of the bivalve mollusk Dreissena polymorpha with the ciliate Conchophthirus acuminatus in three lakes of Belarus (Mastitsky S.E. // BioInvasions Records.
2012. V. 1. P 161–169). In the table with the initial data, which we download from the figshare website, we will be interested in two variables: the length of the mollusk shell (ZMlength, mm) and the number of ciliates found in the mollusk (CAnumber). This example will be discussed in detail in Chapters 4 and 5, so here we will not dwell in detail on the meaning of the analysis, but will focus on the technique of working with Rcmdr.
Next, we define the data loading mode and the Internet link address in the pop-up windows. It is easy to see that we could easily download the same data from the local text file, Excel workbooks or database tables. To make sure that our data is loaded correctly (or edit it if necessary), click the “View data” button.
Window for defining data organization Fragment of the loaded table
At the second stage, in the “Statistics” menu, select “Correlation test”:
We select a pair of correlated variables and in the Output Window we obtain the Pearson correlation coefficient (R = 0.467), the level of statistical significance achieved (p-value 2.2e-16) and 95% confidence limits.
– – –
The results obtained can be easily copied from the output window via the clipboard.
Now we get a graphical representation of the correlation dependence. Let's select a scatterplot of the dependence of CAnumber on ZMlength and provide it with edge range diagrams, a linear trend line using the least squares method (in green), a line smoothed using the local regression method (in red), presented with a confidence region (dotted line). For each of the three lakes (Lake variable), the experimental points will be represented by different symbols.
– – –
Graph copied from the R Commander graphics window As the equivalent of all the R Commander menu button clicks, R language instructions appear in the script window.
In our case they look like this:
Shellfish read.table("http://figshare.com/media/download/98923/97987", header=TRUE, sep="\t", na.strings="NA", dec=".", strip. white=TRUE) cor.test(Clams$CAnumber, Clams$ZMlength, alternative="two.sided", method="pearson") scatterplot(CAnumber ~ ZMlength | Lake, reg.line=lm, smooth=TRUE, spread= TRUE, boxplots="xy", span=0.5, ylab="Number of ciliates", xlab="Shell length", by.groups=FALSE, data=Mollusks) The script itself or the output results (as well as both together ) can be saved in files and repeated at any time. You can get the same result without running R Commander by loading the saved file through the R console.
By and large, without knowing the constructs of the R language (or simply not wanting to burden your memory with remembering them), using Rcmdr you can perform data processing using almost all basic statistical methods. It presents parametric and nonparametric tests, methods for fitting various continuous and discrete distributions, analysis of multivariate contingency tables, univariate and multivariate analysis of variance, principal component analysis and clustering, various forms of generalized regression models, etc. The developed apparatus for analyzing and testing the resulting models is worthy of careful study .
A detailed description of techniques for working with R Commander, as well as features of the implementation of data processing algorithms, can be found in the manuals (Larson-Hall, 2009; Karp, 2014).
However, just as sign language cannot replace human communication in natural language, knowledge of the R language significantly expands the user's capabilities and makes communication with the R environment enjoyable and exciting. And here automatic generation Scripting in R Commander can be an excellent way for the reader to become familiar with R language operators and learn the specifics of calling individual functions. We will devote subsequent chapters of the manual to a discussion of data processing procedures only at the level of language constructs.
1.4. Objects, packages, functions, devices The R language belongs to the family of so-called high-level object-oriented programming languages. For a non-specialist, a strict definition of the concept “object” is quite abstract. However, for simplicity, we can call everything that was created while working with R objects.
There are two main types of objects:
1. Objects intended for storing data (“data objects”) are individual variables, vectors, matrices and arrays, lists, factors, data tables;
2. Functions (“function objects”) are named programs designed to create new objects or perform certain actions on them.
Objects of the R environment intended for collective and free use, are packaged into packages united by similar topics or data processing methods. There is some difference between the terms package ("package") and library ("library"). The term "library" defines a directory that can contain one or more packages. The term "package" refers to a collection of functions, HTML manual pages, and example data objects intended for testing or training purposes.
Packages are installed in a specific directory of the operating system or, in an uninstalled form, can be stored and distributed in archived *. zip files Windows (the package version must match the specific version of your R).
Full information about the package (version, main thematic area, authors, dates of changes, licenses, other functionally related packages, a complete list of functions indicating their purpose, etc.) can be obtained by the command
library(help=package_name), for example:
library(help=Matrix) All R packages fall into one of three categories: basic ("base"), recommended ("recommended") and other user-installed.
You can get a list of them on a specific computer by issuing the library() command or:
installed.packages(priority = "base") installed.packages(priority = "recommended") # Getting full list packages packlist - rownames(installed.packages()) # Output information to the clipboard in Excel format write.table(packlist,"clipboard",sep="\t", col.names=NA) Basic and recommended packages are usually included to the R installation file.
Of course, there is no need to immediately install many different packages in reserve.
To install a package, simply select the “Packages Install package(s)” menu item in the R Console command window or enter, for example, the command:
install.packages(c("vegan", "xlsReadWrite", "car"))
Packages can be downloaded, for example, from the Russian “mirror” http://cran.gis-lab.info, for which it is convenient to use the edition of the Rprofile.site file as shown in section 1.1.
Another option for installing packages is to go to the website http://cran.gis-lab.info/web/packages, select the desired package as a zip file and download to the selected folder on your computer.
In this case, you can preview all the information on the package, in particular, a description of the functions included in it, and decide how much you need it. Next, you need to execute the command menu item “Packages Install packages from local zip files”.
When you start the RGui console, only some core packages are loaded. To initialize any other package, you must issue the library (package_name) command before using its functions directly.
You can determine which packages are loaded at each moment of the session by issuing the command:
sessionInfo() R version 2.13.2 (2011-09-30) Platform: i386-pc-mingw32/i386 (32-bit)
– – –
other attached packages:
Vegan_2.0-2 permute_0.6-3
loaded via a namespace (and not attached):
Grid_2.13.2 lattice_0.19-33 tools_2.13.2 We provide in the following table a list (perhaps not exhaustively complete) of packages that were used in the scripts presented in this book:
R packages Purpose "Basic" packages Basic constructs R base Package compiler R compiler A set of tables with data for testing and demonstrating functions datasets Basic graphics functions graphics Graphics device drivers, color palettes, fonts grDevices Functions for creating graphic layers grid Object-oriented programming components (classes , methods methods) Functions for working with regression splines of various types splines Basic functions of statistical analysis stats Methods of statistical functions of the S4 class stats4 Components of the user interface (menus, selection boxes, etc.) tcltk Information support, administration and documentation tools Various debugging, input utilities output, archiving, etc.
Utils "Recommended" packages Functions of various bootstrap and jackknife procedures boot Various algorithms for non-hierarchical classification and recognition class Algorithms for partitioning and hierarchical clustering cluster Analysis and verification of codes R codetools Reading and writing files in various formats (DBF, SPSS, DTA, Stata) foreign Functions supporting optimization of kernel smoothing KernSmooth Graphical functions of extended functionality (Sarkar, 2008) lattice Set of data and statistical functions (Venables, Ripley, 2002) MASS Operations with matrices and vectors Matrix Generalized additive and mixed effects models mgcv Linear and nonlinear models with mixed effects nlme Neural networks direct propagation nnet Construction of classification and regression trees rpart Functions of kriging and analysis of the spatial distribution of points spatial Survival analysis (Cox model, etc.) survival Packages installed during the work adegenet Algorithms for analyzing genetic distances arm Analysis of regression models - appendix to the book (Gelman, Hill , 2007) car Procedures related to applied regression analysis corrplot Display of correlation matrices in graphical form fitdistrplus Selection of parameters of statistical distributions FWDselect, Selection of a set of informative variables in regression models packfor gamair Data sets for testing additive geosphere models Estimation of geographic distances ggplot2 Advanced graphics package with high functionality DAAG Data analysis and graphics functions for the book (Maindonald, Braun, 2010) Hmisc Harrell's set of functions HSAUR2 Supplement to the book (Everitt, Hothorn, 2010) ISwR Primary statistical analysis in R jpeg Working with graphics jpeg files lars Special types of regression (LARS, Lasso, etc.) lavaan Confirmatory analysis and structural equation models lmodel2 Implementation of regression models of types I and II (MA, SMA, RMA) maptools Tools for working with geographical maps mice Procedures for analyzing and filling in missing values moments Functions for calculating sample moments nortest Criteria for testing the hypothesis of a normal distribution outliers Analysis of outliers in data pastecs Analysis of spatial and time series in ecology pls Regression on principal components pwr Estimation of the statistical power of hypotheses reshape Flexible transformation of robustbase data tables Robust methods for constructing regression models rootSolve Finding the roots of a function with several variables scales Selection of color scales sem Structural equation models semPlot Visualization of structural relationships sm Estimation of distribution density and smoothing methods sp Classes and methods for accessing spatial data spatstat Methods of spatial statistics, selection of models spdep Spatial dependencies: geostatistical stargazer methods and modeling Outputting information about statistical models in different vcd formats Visualizing categorical data Performing calculations on community ecology (measures of similarity, diversity and vegan nesting, ordination and multivariate analysis) If we try to load a package not yet installed in R, or try to use functions of a package that has not yet been loaded, we will receive system messages:
sem(model, data=PoliticalDemocracy) Error: can't find function "sem" library(lavaan) Error in library(lavaan) : no package called "lavaan" The following function, introduced by K. Cichini, takes as an input a list of used user of packages and figures out which ones should be downloaded and which ones need to be pre-installed. Understanding the script requires knowledge of the R language constructs described in the next section, but the interested reader can return to these commands later.
instant_pkgs - function(pkgs) ( pkgs_miss - pkgs)] # Install packages that are not ready for download:
if (length(pkgs_miss) 0) ( install.packages(pkgs_miss) ) # Download packages that have not yet been downloaded:
Attached - search() attached_pkgs - attached need_to_attach - pkgs if (length(need_to_attach) 0) ( for (i in 1:length(need_to_attach)) require(need_to_attach[i], character.only = TRUE) ) ) # Call example:
instant_pkgs(c("base", "jpeg", "vegan"))
You can get a list of the functions of each package, for example, by running the command:
ls(pos = "package:vegan") Note: ls() is a general purpose function for listing objects in a given environment. The command above installs the vegan package as such an environment. If we issue this command without parameters, we will get a list of objects created during the current session.
You can get a list of arguments to the incoming parameters of any function in a loaded package by issuing the args() command.
For example, when running the linear model obtaining function lm(), which we later widely use, the parameters are set:
Args(lm) function (formula, data, subset, weights, na.action, method = "qr", model = TRUE, x = FALSE, y = FALSE, qr = TRUE, singular.ok = TRUE, contrasts = NULL, offset,...) If you enter a command consisting only of an abbreviation of a function (for example, calculating the interquartile range of IQR), you can get the source text of the function in R language codes:
IQR function (x, na.rm = FALSE) diff(quantile(as.numeric(x), c(0.25, 0.75), na.rm = na.rm, names = FALSE)) An advanced user can make changes to this code and “redirect” the standard function call to your version.
However, if we want to look at the code of the predict() function, which is used to calculate the predicted values of the linear model, in the same way, we will get:
predict function (object,...) UseMethod("predict") In this case, predict() is a "universal" function: depending on which model object is fed to its input (lm for linear regression, glm for Poisson or logistic regression, lme for mixed effects model, etc.), the appropriate method for obtaining predicted values is updated.
Specifically, this function is used to implement the following methods:
methods("predict") predict.ar* predict.Arima* predict.arima0* predict.glm predict.HoltWinters* predict.lm predict.loess* predict.mlm predict.nls* predict.poly predict.ppr* predict.prcomp* predict.princomp* predict.smooth.spline* predict.smooth.spline.fit* predict.StructTS* Non-visible functions are asterisked This example relates to the object-oriented programming (OOP) ideas underlying the R framework. For OOP in In S3 style, a method is, strictly speaking, a function that is called by another generic function, such as print(), plot() or summary(), depending on the class of the object supplied to its input. In this case, the class attribute is responsible for “object orientation,” which ensures correct dispatch and calling the required method for a given object. So the “method function” for obtaining predicted values of a generalized linear model will have a call to predict.glm(), when smoothing with splines – predict.smooth.spline(), etc. Detailed information about the S3 OOP model can be found in the S3Methods help section, and for the more advanced S4 model, in the Methods section.
Finally, let's look at some simple techniques for saving work produced during an R session:
° sink(file= file name) – outputs the results of executing subsequent commands in real time to a file with the given name; To terminate this command, you must run the sink() command without parameters;
° save(file= file name, list of saved objects) – saves the specified objects in a binary file of XDR format, which can be worked with in any operating system;
° load(file= file name) – restores saved objects in the current environment;
° save.image(file= file name) – saves all objects created during the work as an R-specific rda file.
An example of transferring a generated table with data to the clipboard in a format compatible with the Excel sheet structure was given above in this section. Chapter 6 will provide an example of transferring data from a linear model object to a Word file.
The R environment can generate a pixel image required quality for almost any display resolution or printing device, and also save the resulting graphic windows in files of different formats. There is a driver function for each graphics output device: you can enter the help(Devices) command to get a complete list of drivers.
Among the graphics devices, the most common are:
° windows() – graphical Windows window(screen, printer or metafile).
° png(), jpeg(), bmp(), tiff() – output to a raster file of the appropriate format;
° pdf(),postscript() – output of graphic information in PDF file or PostScript.
When you are finished working with the output device, you should disable its driver using the dev.off() command. It is possible to activate several graphic output devices simultaneously and switch between them: see, for example, the corresponding section in the book by Shipunov et al. (2012, p. 278).
1. DESCRIPTION OF THE R LANGUAGE
2.1. Data types of the R language All data objects (and therefore variables) in R can be divided into the following classes (i.e. object types):
° numeric – objects that include integers (integer) and real numbers (double);
° logical – logical objects that take only two values: FALSE (abbreviated F) and TRUE (T);
° character – character objects (variable values are specified in double or single quotes).
In R, you can create names for various objects (functions or variables) in both Latin and Cyrillic, but please note that a (Cyrillic) and a (Latin) are two different objects. Additionally, the R environment is case sensitive, i.e. lowercase and capital letters are different. Variable names (identifiers) in R must begin with a letter (or period) and consist of letters, numbers, periods and underscores.
With the help of a team? name, you can check whether a variable or function with the specified name exists.
Checking whether a variable belongs to a specific class is checked by the functions is.numeric(object_name), is.integer(name), is.logical(name), is.character(name), and to convert an object to another type you can use the functions as.numeric (name), as.integer(name), as.logical(name), as.character(name).
There are a number of special objects in R:
° Inf – positive or negative infinity (usually the result of dividing a real number by 0);
° NA – “missing value” (Not Available);
° NaN – “not a number”.
You can check whether a variable belongs to any of these special types using the functions is.nite(name), is.na(name) and is.nan(name), respectively.
An R expression is a combination of elements such as an assignment operator, arithmetic or logical operators, object names, and function names. The result of executing an expression is usually immediately displayed in the command or graphics window. However, when an assignment operation is performed, the result is stored in the corresponding object and is not displayed on the screen.
As an assignment operator in R, you can use either the “=” symbol, or a pair of symbols “-“ (assigning a specific value to the object on the left) or “-“ (assigning a value to the object on the right). It is considered good programming style to use “-“.
R language expressions are organized line by line in a script. You can enter several commands on one line, separating them with the symbol “;“. One command can also be placed on two (or more) lines.
numeric objects can form expressions using traditional arithmetic operations + (addition), – (subtraction), * (multiplication), / (division), ^ (exponentiation), %/% (integer division), %% (remainder) from division). Operations have normal priority, i.e. First, exponentiation is performed, then multiplication or division, then addition or subtraction. Expressions can use parentheses and operations within them have the highest priority.
Boolean expressions can be composed using the following logical operators:
° "Equal to" == ° "Not equal to" != ° "Less than" ° "Greater than" ° "Less than or equal to" = ° "Greater than or equal to" = ° "Logical AND" & ° "Logical OR" | ° "Logical NOT" !
SUPPORT, AU TSOURCING SERVICES G&A FUNDS ADMINISTRATION 2nd AMICORP GROUP COMPANY'S AREA OF OPERATION STAND OUT FROM THE CROWD w w w.am icor p. c om AMICORP GROUP FIELD OF OPERATIONS OF THE COMPANY CONTENTS ABOUT THE COMPANY OUR US MEADOWS Services for corporate clients Services for institutional sales Creation and management...”
“Federal State Educational Budgetary Institution of Higher Professional Education “Financial University under the Government of the Russian Federation” Department of “Marketing” MODERN DIRECTIONS OF MARKETING: THEORY, METHODOLOGY, PRACTICE COLLECTIVE MONOGRAPHY Under the general editorship of S.V. Karpova Moscow 2011 Reviewers: N.S. Perekalina - Doctor of Economics, Professor, Head. Department of Marketing "MATI" - Russian State Technological University named after. K. E. Tsiolkovsky S.S. Soloviev..."
“Little digital newsletter for CAFE and TEA RDACTION: Ch. Editor: Vesela Dabova Br.4 December, 2011 Editors: Otslabvane s chai Teodora Vasileva Gergana Ivanov Publishing house: ABB How are all the cases in the body when taking the tea drink and how are all the cases weakened by Nai-sigurniyat and healthy beginnings for namalyavane on tegloto e redovnata consummation for a cup of tea. There are different opinions regarding the reliability of the theory, but very few facts prove that every cup of tea is combined with the defined regime on...”
"INTERNATIONAL INTERDISCIPLINARY SCIENTIFIC CONFERENCE RADICAL SPACE IN BETWEEN DISCIPLINES RCS 2015 CONFERENCE BOOK OF ABSTRACTS EDITORS Romana Bokovi Miljana Zekovi Slaana Milievi NOVI SAD / SERBIA / SEPTEMBER 21-23 / 2015 Radical Space In Between Disciplines Conference Book of Abstracts Editors: a Bokovi Miljana Zekovi Slaana Milievi ISBN: 978-86-7892-733-1 Layout: Maja Momirov Cover design: Stefan Vuji Published by Department of Architecture and Urbanism, Faculty of Technical Sciences,...”
“SAINT PETERSBURG STATE UNIVERSITY Faculty of Geography and Geoecology Department of Geomorphology DIPLOMA THESIS (final qualifying work) on the topic: “Geomorphological features and paleoclimate of Arctic lakes (using the example of lakes in the central sector of the Russian Arctic)” Completed by: evening student Elena Aleksandrovna Morozova Scientific advisors: Doctor of Geography, Prof. Bolshiyanov Dmitry Yurievich Ph.D., senior teacher Savelyeva Larisa Anatolyevna Reviewer: Ph.D., head...”
“Apacer M811 mouse is a laser mini-SUV for the Kit. http://news.kosht.com/computer/mouse/2009/11/26/mysh_apacer_m811. search plugin for daily prices KOSHT.com for Firefox browser. Install One Click. One kilobyte. Home News Prices Announcements Jobs Forums Companies Mobi Find Find your news All KOSTA news PCs and components Mice PCs and components Mice All KOSTA news Best gaming computers On-line calculation at UltraPrice.by Mouse Apacer M811 – laser mini-SUV [...»
"FEDERAL EDUCATIONAL AGENCY STATE EDUCATIONAL INSTITUTION OF HIGHER PROFESSIONAL EDUCATION MOSCOW STATE INDUSTRIAL UNIVERSITY (GOU MGIU) "INFORMATION SYSTEMS AND TECHNOLOGIES" DEPARTMENT OF DEGREE THESIS majoring in "Mathematical support and administration" information systems» student Tatyana Andrevna Chumakova on the topic “Calculation of separated flows behind a poorly streamlined body” Head of work: Prof., Doctor of Physics and Mathematics. n. Aleksin Vladimir Adamovich..."
“R WIPO A/45/3 ORIGINAL: English DATE: August 15, 2008 WORLD INTELLECTUAL PROPERTY ORGANIZATION GENEVA ASSEMBLY OF MEMBER STATES WIPO Forty-fifth Series of Meetings Geneva, September 22-30, 2008 ADMISSION OF OBSERVERS General Director I. ADMISSION OF INTERNATIONAL NON-GOVERNMENTAL ORGANIZATIONS AS OBSERVERS 1. At their previous sessions, the Assemblies adopted a number of principles to be applied when referring international non-governmental organizations...”
“1 Oleg Sanaev. A ROUND THE WORLD COURSE THAT LASTS FOUR YEARS AND COSTS A HUNDRED DOLLARS With the duration of Evgeniy Aleksandrovich Gvozdev’s trip on the yacht Lena, indicated in the title, everything is in order - four years plus two weeks: on July 7, 1992, he left the Makhachkala port, on July 19, 1996, he returned . But with money, it’s a clear exaggeration, or rather an understatement: you can’t, of course, live on a hundred dollars for four years - you’ll stretch your legs. But when starting his voyage, Gvozdev had exactly this amount at his disposal. And at least the legs..."
"Institute of Management, Research University Belgorod State National Research University TECHNOLOGIES OF SECURITY FORMATION SECURING THE FORMATION OF PERSONNEL RESERVE CANDIDATE POOL STATE FOR STATE AND MUNICIPAL AND MUNICIPAL SERVICE Abstract: Summary: The article discusses..."
“Lydia YANOVSKAYA NOTES ABOUT MIKHAIL BULGAKOV MOSCOW “TEXT” UDC 821.161.1 BBK 84 (2Ros-Rus)6-44 Ya64 ISBN 978-5-7516-0660-2 ISBN 978-985-16-3297-4 (Harvest LLC ") "Text", 2007 "BRAVO, BIS, PAWNSHOP!" “BRAVO, BIS, PAWNSHOP!” I don’t know where the editorial office of the Yunost magazine is located in Moscow today. Does such a magazine still exist? In the mid-70s, this youngest and prettiest editorial office in Moscow was located on Sadovaya-Triumfalnaya, next to Mayakovsky Square, occupying a small but extremely cozy..."
“Appendix 1 APPLICATION FORMS FOR COMPETITIONS 2013 Form “T”. Title page of the application to the Russian Humanitarian Science Foundation Project name Project number Project type (a, c, d, e, f) Area of knowledge (code) Russian Humanitarian Science Foundation classifier code GRNTI code (http://www.grnti.ru/) Priority direction of development of science and technology and technology in the Russian Federation, critical technology1 Last name, first name, patronymic of the manager Contact telephone number of the project manager of the project Full and short name of the organization through which it should be carried out...”
“FNI Report 8/2014 Implementing EU Climate and Energy Policies in Poland: From Europeanization to Polonization? Jon Birger Skjrseth Implementing EU Climate and Energy Policies in Poland: From Europeanization to Polonization? Jon Birger Skjrseth [email protected] December 2014 Copyright © Fridtjof Nansen Institute 2014 Title Implementing EU Climate and Energy Policies in Poland: From Europeanization to Polonization? Publication Type and Number Pages FNI Report 8/2014 57 Author ISBN 978-82-7613-683-8 Jon...”
""Scientific notes of TOGU" Volume 6, No. 4, 2015 ISSN 2079-8490 Electronic scientific publication "Scientific notes of TOGU" 2015, Volume 6, No. 4, P. 173 - 178 Certificate El No. FS 77-39676 dated 05.05.2010 http http://pnu.edu.ru/ru/ejournal/about/ [email protected] UDC 316.33 © 2015 I. A. Gareeva, Doctor of Sociology. Sciences, A. G. Kiseleva (Pacific State University, Khabarovsk) FORMATION OF SOCIAL INSURANCE SYSTEMS This article analyzes the formation of social insurance systems and its current state...”
“Conference Program Chiang Mai, Thailand November, 2015 APCBSS Asia -Pacific Conference on Business & Social Sciences ICEI International Conference on Education Innovation APCLSE Asia-Pacific Conference on Life Science and Engineering APCBSS Asia -Pacific Conference on Business & Social Sciences ISBN978-986- 90263-0-7 ICEI International Conference on Education Innovation ISBN 978-986-5654-33-7 APCLSE Asia-Pacific Conference on Life Science and Engineering ISBN 978-986-90052-9-6 Content Content..."
Last time (in November 2014; I’m very ashamed that I took so long with the continuation!) I talked about the basic capabilities of the R language. Despite the presence of all the usual control constructs, such as loops and conditional blocks, the classical approach to data processing iteration-based is far from the best solution, since loops in R unusually slow. So now I’ll tell you how you actually need to work with data so that the calculation process doesn’t force you to drink too many cups of coffee waiting for the result. In addition, I will spend some time talking about how to use modern means data visualization in R. Because the convenience of presenting data processing results in practice is no less important than the results themselves. Let's start with something simple.
Vector operations
As we remember, the basic type in R is not a number at all, but a vector, and the basic arithmetic operations operate on vectors element-by-element:
> x<- 1:6; y <- 11:17 >x + y 12 14 16 18 20 22 18 > x > 2 FALSE FALSE TRUE TRUE TRUE TRUE > x * y 11 24 39 56 75 96 17 > x / y 0.09090909 0.16666667 0.23076923 0.28571429 0.3 3333333 0.37500000 0.05882353
Everything here is quite simple, but it is quite logical to ask the question: what will happen if the lengths of the vectors do not match? If we, say, write k<- 2, то будет ли x * k соответствовать умножению вектора на число в математическом смысле? Короткий ответ - да. В более общем случае, когда длина векторов не совпадает, меньший вектор просто продолжается повторением:
> z<- c(1, 0.5) >x * z 1 1 3 2 5 3
The situation is approximately the same with matrices.
> x<- matrix(1:4, 2, 2); y <- matrix(rep(2,4), 2, 2) >x * y [,1] [,2] 2 6 4 8 > x / y [,1] [,2] 0.5 1.5 1.0 2.0
In this case, “normal” and not bitwise matrix multiplication will look like this:
> x %*% y [,1] [,2] 8 8 12 12
All this, of course, is very good, but what do we do when we need to apply our own functions to the elements of vectors or matrices, that is, how can this be done without a loop? The approach that R uses to solve this problem is very similar to what we are used to in functional languages - it is reminiscent of the map function in Python or Haskell.
Useful function lapply and its friends
The first function in this family is lapply. It allows you to apply a given function to each element of a list or vector. Moreover, the result will be exactly the list, regardless of the type of argument. The simplest example using lambda functions:
>q<- lapply(c(1,2,4), function(x) x^2) >q 1 4 16
If the function you want to apply to a list or vector requires more than one argument, then those arguments can be passed through lapply.
>q<- lapply(c(1,2,4), function(x, y) x^2 + y, 3)
The function works in a similar way with a list:
> x<- list(a=rnorm(10), b=1:10) >lapply(x, mean)
Here, the rnorm function specifies the normal distribution (in this case, ten normally distributed numbers ranging from 0 to 1), and mean calculates the average value. The sapply function is exactly the same as the lapply function except that it attempts to simplify the result. For example, if each element of a list is of length 1, then a vector will be returned instead of a list:
> sapply(c(1,2,4), function(x) x^2) 1 4 16
If the result is a list of vectors of the same length, then the function will return a matrix, but if nothing is clear, then just a list, like lapply.
> x<- list(1:4, 5:8) >sapply(x, function(x) x^2) [,1] [,2] 1 25 4 36 9 49 16 64
To work with matrices, it is convenient to use the apply function:
> x<- matrix(rnorm(50), 5, 10) >apply(x, 2, mean) > apply(x, 1, sum)
Here, to begin with, we create a matrix of five rows and ten columns, then first we calculate the average over the columns, and then the sum in the rows. To complete the picture, it should be noted that the task of calculating the average and sum of rows is so common that R provides special functions for this purpose rowSums, rowMeans, colSums and colMeans.
The apply function can also be used for multidimensional arrays:
> arr<- array(rnorm(2 * 2 * 10), c(2, 2, 10)) >apply(arr, c(1,2), mean)
The last call can be replaced with a more readable option:
> rowMeans(arr, dim = 2)
Let's move on to the mapply function, which is a multidimensional analogue of lapply. Let's start with a simple example, which can be found directly in the standard R documentation:
> mapply(rep, 1:4, 4:1) 1 1 1 1 2 2 2 3 3 4
As you can see, what happens here is that the rep function is applied to a set of parameters that are generated from two sequences. The rep function itself simply repeats the first argument the number of times specified as the second argument. So the previous code is simply equivalent to the following:
> list(rep(1,4), rep(2,3), rep(3,2), rep(4,1))
Sometimes it is necessary to apply a function to some part of an array. This can be done using the tapply function. Let's look at the following example:
> x<- c(rnorm(10, 1), runif(10), rnorm(10,2)) >f<- gl(3,10) >tapply(x,f,mean)
First, we create a vector, the parts of which are formed from random variables with different distributions, then we generate a vector of factors, which is nothing more than ten ones, then ten twos and the same number of threes. Then we calculate the average for the corresponding groups. The tapply function by default tries to simplify the result. This option can be disabled by specifying simplify=FALSE as the parameter.
> tapply(x, f, range, simplify=FALSE)
When people talk about the apply functions, they usually also talk about the split function, which splits a vector into pieces, similar to tapply . So, if we call split(x, f) we will get a list of three vectors. So the lapply/split pair works the same as tapply with the simplify value set to FALSE:
> lapply(split(x, f), mean)
The split function is useful beyond working with vectors: it can also be used to work with data frames. Consider the following example (I borrowed it from the R Programming course on Coursera):
> library(datasets) > head(airquality) Ozone Solar.R Wind Temp Month Day 1 41 190 7.4 67 5 1 2 36 118 8.0 72 5 2 3 12 149 12.6 74 5 3 4 18 313 11.5 62 5 4 5 NA NA 14.3 56 5 5 6 28 NA 14.9 66 5 6 > s<- split(airquality, airquality$Month) >lapply(s, function(x) colMeans(x[, c("Ozone", "Solar.R", "Wind")]))
Here we are working with a dataset that contains information about air conditions (ozone, solar radiation, wind, temperature in Fahrenheit, month and day). We can easily report monthly averages using split and lapply as shown in the code. Using sapply, however, will give us the result in a more convenient form:
> sapply(s, function(x) colMeans(x[, c("Ozone", "Solar.R", "Wind")])) 5 6 7 8 9 Ozone NA NA NA NA NA Solar.R NA 190.16667 216.483871 NA 167.4333 Wind 11.62258 10.26667 8.941935 8.793548 10.1800
As you can see, some value values are not defined (and the reserved value NA is used for this). This means that some (at least one) values in the Ozone and Solar.R columns were also undefined. In this sense, the colMeans function behaves completely correctly: if there are any undefined values, then the average is therefore undefined. The problem can be solved by forcing the function to ignore NA values using the na.rm=TRUE parameter:
> sapply(s, function(x) colMeans(x[, c("Ozone", "Solar.R", "Wind")], na.rm=TRUE)) 5 6 7 8 9 Ozone 23.61538 29.44444 59.115385 59.961538 31.44828 Solar.R 181.29630 190.16667 216.483871 171.857143 167.43333 Wind 11.62258 10.26667 8.941935 8.793548 10.18000
Why do you need so many functions to solve very similar problems? I think this question will be asked by every second person who has read all this. All of these functions are actually trying to solve the problem of processing vector data without using loops. But it's one thing to achieve high data processing speed, and quite another to gain at least some of the flexibility and control that control constructs such as loops and conditional statements provide.
Data visualization
The R system is incredibly rich in data visualization tools. And here I am faced with a difficult choice - what to talk about if the area is so large. If in the case of programming there is some basic set of functions, without which nothing can be done, then in visualization there is a huge amount various tasks and each of them (usually) can be solved in several ways, each of which has its own pros and cons. Moreover, there are always many options and packages that allow you to solve these problems in different ways.
About standard means A lot has been written about visualization in R, so here I would like to talk about something more interesting. In recent years, the package has become increasingly popular ggplot2, so let's talk about him.
In order to start working with ggplot2, you need to install the library using the install.package("ggplot2") command. Next, we connect it for use:
> library("ggplot2") > head(diamonds) carat cut color clarity depth table price x y z 1 0.23 Ideal E SI2 61.5 55 326 3.95 3.98 2.43 2 0.21 Premium E SI1 59.8 61 326 3.89 3.84 2.31 3 0.23 Good E VS1 56. 9 65 327 4.05 4.07 2.31 4 0.29 Premium I VS2 62.4 58 334 4.20 4.23 2.63 5 0.31 Good J SI2 63.3 58 335 4.34 4.35 2.75 6 0.24 Very Good J VVS2 62.8 57 336 3.9 4 3.96 2.48 > head(mtcars) mpg cyl disp hp drat wt qsec vs am gear carb Mazda RX4 21.0 6 160 110 3.90 2.620 16.46 0 1 4 4 Mazda RX4 Wag 21.0 6 160 110 3.90 2.875 17.02 0 1 4 4 Datsun 710 22.8 4 108 93 3.85 2. 320 18.61 1 1 4 1 Hornet 4 Drive 21.4 6 258 110 3.08 3.215 19.44 1 0 3 1 Hornet Sportabout 18.7 8 360 175 3.15 3.440 17.02 0 0 3 2 Valiant 18.1 6 225 105 2.76 3.460 20.22 1 0 3 1
The diamonds and mtcars data are part of the ggplot2 package, and it is with them that we will now work. With the first, everything is clear - this is data about diamonds (clarity, color, cost, etc.), and the second set is data from road tests (number of miles per gallon, number of cylinders...) of cars produced in 1973–1974 from the American magazine Motor Trends . More detailed information about the data (for example, dimension) can be obtained by typing ?diamonds or ?mtcars .
For visualization, the package provides many functions, of which qplot will be the most important for us now. The ggplot function gives you significantly more control over the process. Anything that can be done with qplot can also be done with ggplot. Let's look at this simple example:
> qplot(clarity, data=diamonds, fill=cut, geom="bar")
The same effect can be achieved with the ggplot function:
> ggplot(diamonds, aes(clarity, fill=cut)) + geom_bar()
However, calling qplot looks simpler. In Fig. 1 you can see how the dependence of the number of diamonds with different cut qualities on clarity is plotted.
Now let’s plot the dependence of the mileage per unit of fuel of cars on their mass. The resulting scatter plot (or scatter plot scatter plot) presented
in Fig. 2.
> qplot(wt, mpg, data=mtcars)
You can also add color display of the quarter mile acceleration time (qsec):
> qplot(wt, mpg, data=mtcars, color=qsec)

When visualizing, you can also transform data:
> qplot(log(wt), mpg - 10, data=mtcars)
In some cases, discrete color divisions appear more representative than continuous ones. For example, if we want to display in color information about the number of cylinders instead of acceleration time, then we need to indicate that the value is discrete in nature (Fig. 3):
> qplot(wt, mpg, data=mtcars, color=factor(cyl))

You can also change the size of the points using, for example, size=3 . If you are going to print graphs on a black and white printer, then it is better not to use colors, but instead change the shape of the marker depending on the factor. This can be done by replacing color=factor(cyl) with shape=factor(cyl) .
The plot type is specified using the geom parameter, and in the case of scatter plots, the value of this parameter is "points" .
Now let’s say we just want to build a histogram of the number of cars with the corresponding cylinder value:
> qplot(factor(cyl), data=mtcars, geom="bar") > qplot(factor(cyl), data=mtcars, geom="bar", color=factor(cyl)) > qplot(factor(cyl) , data=mtcars, geom="bar", fill=factor(cyl))
The first call simply draws three histograms for different cylinder values. It must be said that the first attempt to add color to the histogram will not lead to the expected result - the black bars will still be black, but will only have a colored outline. But the last call to qplot will produce a beautiful histogram, as shown in Fig. 4.
We should be clear here. The fact is that the current object we have built is not a histogram in the strict sense of the word. Typically, a histogram is a similar display for continuous data. In English bar chart(that's what we just did) and histogram- these are two different concepts (see the corresponding articles on Wikipedia). Here I will, with some heaviness, use the word “histogram” for both concepts, believing that the very nature of the data speaks for itself.

If we return to Fig. 1, then ggplot2 provides several useful options for positioning plots (the default is position="stack"):
> qplot(clarity, data=diamonds, geom="bar", fill=cut, position="dodge") > qplot(clarity, data=diamonds, geom="bar", fill=cut, position="fill") > qplot(clarity, data=diamonds, geom="bar", fill=cut, position="identity")
The first of the proposed options plots diagrams side by side, as shown in Fig. 5, the second shows the proportions of diamonds of different cut qualities in total number diamonds of a given clarity (Fig. 6).


Now let's look at an example of a real histogram:
> qplot(carat, data=diamonds, geom="histogram", bandwidth=0.1) > qplot(carat, data=diamonds, geom="histogram", bandwidth=0.05)
Here the bandwidth parameter just shows how wide the band is in the histogram. A histogram shows how much data is in what range. The results are presented in Fig. 7 and 8.


Sometimes when we need to plot a model (linear or say polynomial), we can do it directly in qplot and see the result. For example, we can plot mpg versus mass wt directly on top of the scatter plot:
> qplot(wt, mpg, data=mtcars, geom=c("point", "smooth"))
By default, local polynomial regression (method="loess") will be used as the model. The result of the work will look as shown in Fig. 9, where the dark gray bar is the standard error. It is displayed by default, you can turn it off by writing se=FALSE .

If we want to try to fit a linear model on this data, then this can be done by simply specifying method=lm (Fig. 10).

And finally, of course, we need to show how to build pie charts:
>t<- ggplot(mtcars, aes(x=factor(1), fill=factor(cyl))) + geom_bar(width=1) >t + coord_polar(theta="y")
Here we'll use the more flexible ggplot function. It works like this: first we build a graph showing the shares of cars with different numbers of cylinders in the total mass (Fig. 11), then we convert the graph to polar coordinates (Fig. 12).


Instead of a conclusion
Now we're comfortable using R. What's next? It is clear that the most basic capabilities of ggplot2 are given here and issues related to vectorization are discussed. There are several good books on R that are worth mentioning, and they are certainly worth consulting more often than the services of a corporation of very intrusive goodness. Firstly, this is the book by Norman Matloff (Norman Matloff) The Art of R Programming. If you already have experience in programming in R, then The R Inferno, written by P. Burns, will be useful to you. The classic book Software for Data Analysis by John Chambers is also quite appropriate.
If we talk about visualization in R, there is a good book R Graphics Cookbook by W. Chang (Winston Chang). The examples for ggplot2 in this article were taken from Tutorial: ggplot2. See you in the next article: Data Analysis and Machine Learning in R!
 Order 343 mail. Order by Russian post. Consequences of failure to appear in court when summoned
Order 343 mail. Order by Russian post. Consequences of failure to appear in court when summoned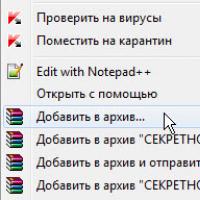 How to put a password on a folder on a Windows computer without and with programs
How to put a password on a folder on a Windows computer without and with programs Pluton – Free Bootstrap HTML5 One Page Template
Pluton – Free Bootstrap HTML5 One Page Template History of ZX Spectrum: Myths and reality New Spectrum
History of ZX Spectrum: Myths and reality New Spectrum Voice assistant Siri from Apple Siri functions on iPhone 6s
Voice assistant Siri from Apple Siri functions on iPhone 6s How to roll back to a previous version of iOS?
How to roll back to a previous version of iOS? Unlock iPad in four days
Unlock iPad in four days